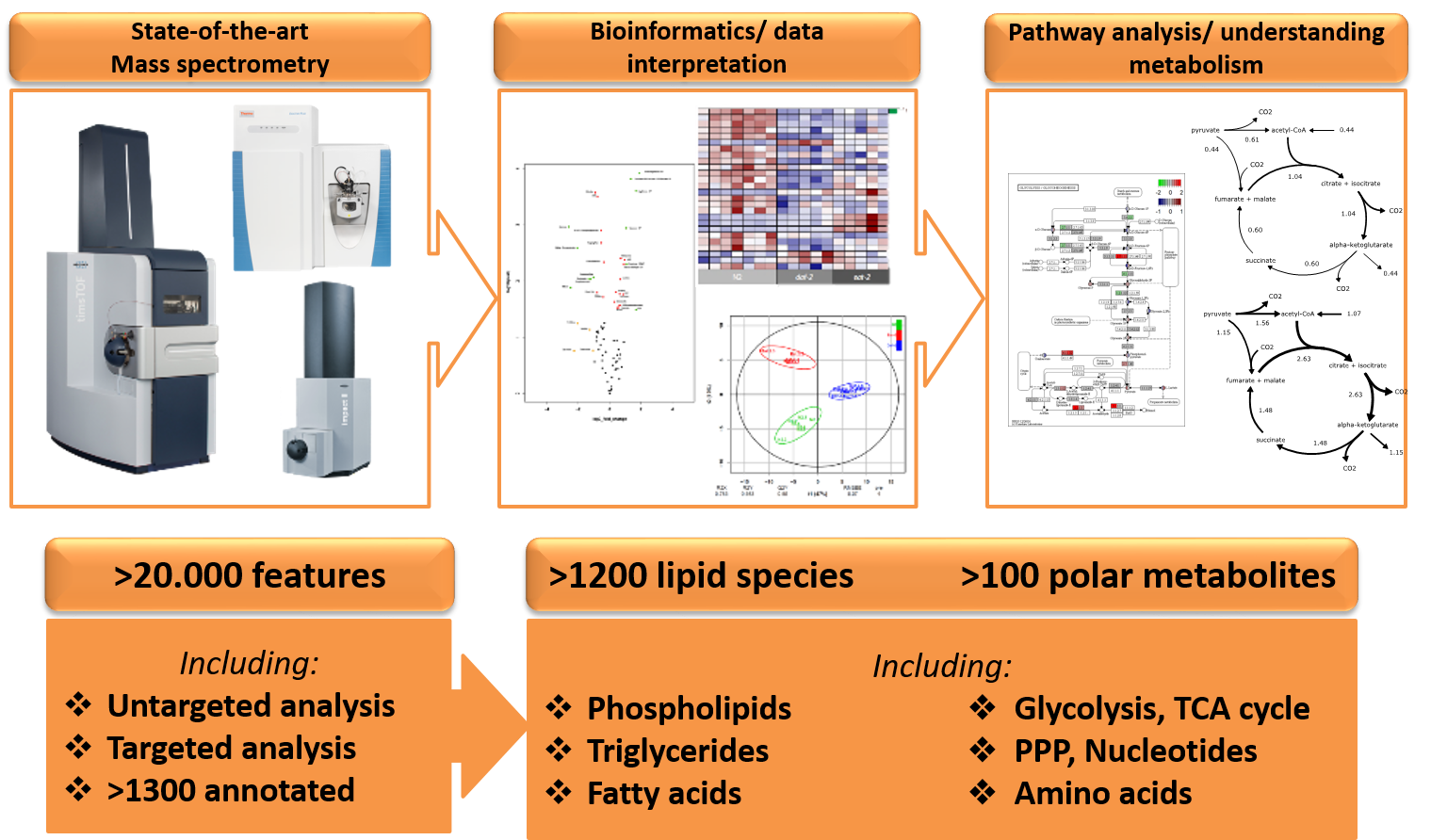
Metabolomics involves the (semi-)quantitative and qualitative measurement of metabolites in biological matrices, including cells, tissues and body fluids. This growing and complex discipline is needed to address the functional questions that arise from the analysis and interpretation of data generated by next-generation sequencing and shot-gun proteomics, techniques that are increasingly used in biomedical research. As the metabolome is most proximal to the phenotype/function it provides a readout for the unique chemical fingerprints that specific cellular processes leave behind. Therefore, metabolomics is the perfect tool to gain insight into (patho)physiological mechanisms of complex systems and is increasingly applied in diverse research fields for biomarker discovery, ranging from oncology, immunology and metabolism. Despite the success of metabolomics in phenotype characterization and biomarker discovery, there still remain conceptual challenges and technical limitations. For example, the use of metabolomics only displays the phenotypical output in a static environment (steady-state differences) and is not able to distinguish the actual dynamics within a cell. Metabolomics after incubation with stable isotope labelled metabolites (i.e. non-radioactive) is expanding analytical possibilities; the time-dependent conversion of stable isotope tracers into downstream products allows the quantification of metabolic pathways (fluxomics), which is closest to metabolic activity in cells or tissues (Figure 1). Using mass isotopomer distribution analysis (MIDA), these stable isotope tracers can be distinguished from their natural abundant occurrence in metabolites. Furthermore, as stable isotopes can safely be applied in animal and human studies, fluxomics experiments are ideal to characterize metabolism in complex systems. This is now routinely applied for small molecule metabolomics to trace glucose metabolism for example, but also increasingly applied for lipidomics to trace the metabolism of these complex biomolecules.
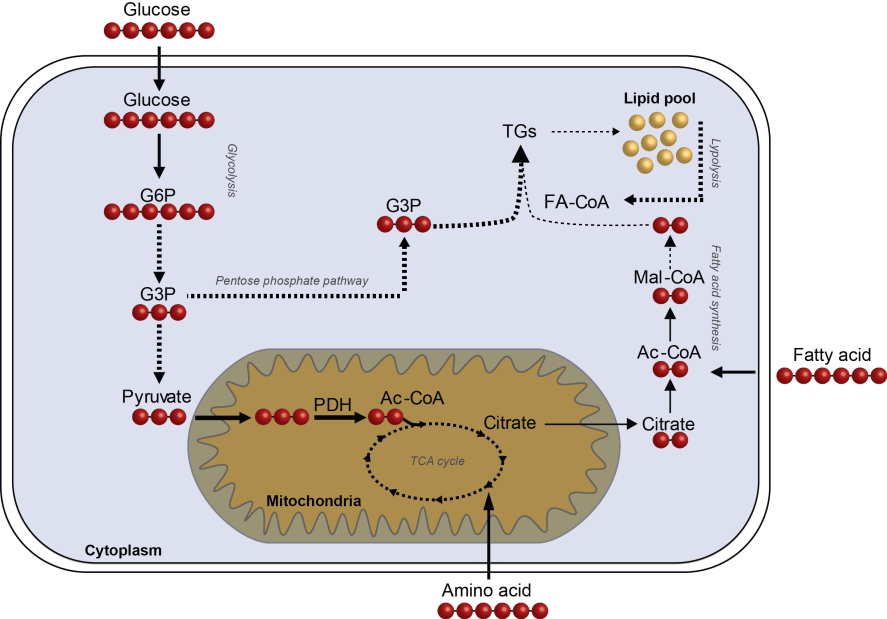
Figure 1. Schematic overview of fluxomics of the central carbon metabolism - The incorporation of 13C-labeled substrates into intermediate metabolites to determine metabolic fluxes in central carbon metabolism, making use of mass isotopomer distribution analysis (MIDA). The red dots indicate the 13C-label which is introduced into the pathway via either glucose, amino acids or fatty acids. In this way, metabolic flexibility and substrate specificity can be determined.